JPhyloIO Simple Alignment Demo
This example application demonstrates how to read multiple sequence alignments into a simple application business model and write them to an output file using JPhyloIO. It is also explained why data adapters are necessary and how they are used for writing.
[Download executable demo files] [Browse source codes]
Overview
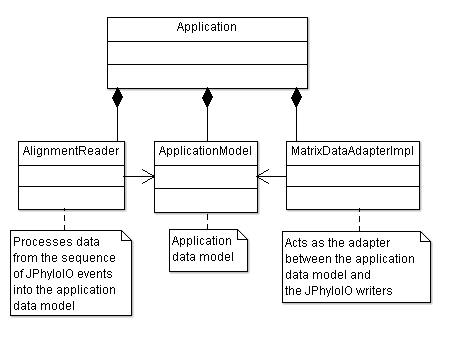
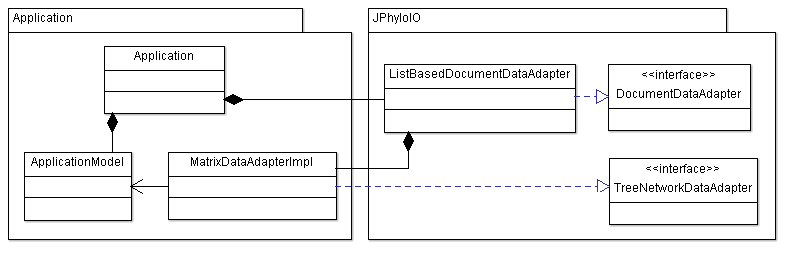
The main class of this example is Application. It offers functionality for reading, displaying and writing multiple sequence alignments. Data can be displayed by an output to the command line. As shown in figure 1 additional classes for reading and writing are part of the application and connect its business model with the event readers and writers of JPhyloIO.
Reading
The main class Application uses AlignmentReader to process the contents of a sequence of JPhyloIOEvents (produced by a class of the type JPhyloIOEventReader) into its business model. A single implementation of such a reader processing the event contents is enough to support all formats included in JPhyloIO. AlignmentReader has a number of methods, each processing the events that correspond to a grammar node in the JPhyloIO event grammar. These events are nested between a start and an end version of an event as defined by its EventTopologyType.
Events of unsupported types and all events nested under them are skipped. Depending on the task the application performs, different types of events may be supported. In case of this example all events containing information about multiple sequence alignments are processed.
Writing
Implementations of the data adapter interfaces (either custom or provided by JPhyloIO) are necessary to provide a JPhyloIOEventWriter with the information necessary for writing. Since different formats may need this information in a different order, it is not possible to write from an event stream without buffering large amounts of data, if the order does not fit the current format. Using data adapters, writers can obtain the data in any order needed, which makes writing more memory efficient.
Data adapter interfaces representing different parts of a phylogenetic document are available in JPhyloIO. An implementation of DocumentDataAdapter provided by JPhyloIO used in this example is ListBasedDocumentDataAdapter. No application specific implementation is necessary here since neither any other element of a document (an OTU list or a phylogenetic tree) nor additional multiple sequence alignments are modeled by the application. The list based document adapter is just filled with a single MatrixDataAdapter.
The custom application data adapter implementation MatrixDataAdapterImpl is necessary to provide direct access to the data modeled in the application business model. In this case it provides access to the sequence IDs and token lists. Sequence names are returned in form of a default prefix with the index of the sequence in question attached to it, e.g. "Sequence 1".