JPhyloIO demos
To understand how JPhyloIO works and can be used, a number of example applications are provided here. The source codes of all these applications can be viewed and downloaded from the subversion repository.
Available examples
Simple alignment demo
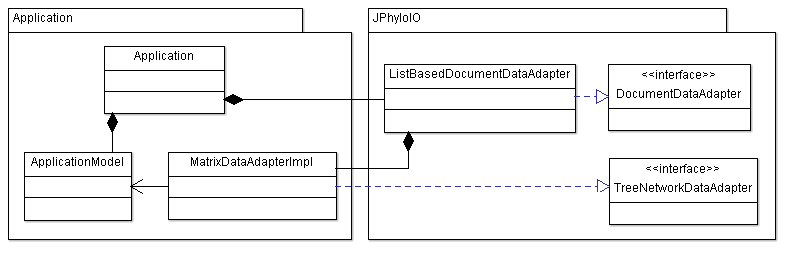
Shows the basic principles of reading and writing data with JPhyloIO and should be used to get started. An application reader and a matrix data adapter are implemented to link an example business model to JPhyloIO.
- Detailed description
- Browse source codes of this demo
- SVN checkout:
https://secure.bioinfweb.info/Code/svn/JPhyloIO/trunk/demo/info.bioinfweb.jphyloio.demo.simplealignment - Download executable demo files
Tree demo
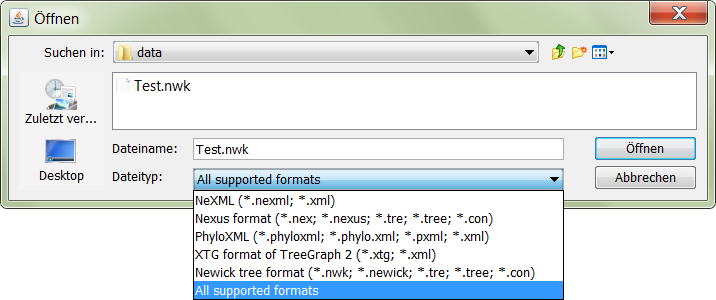
Shows how to read phylogenetic trees from the non-hierarchical tree representation of the JPhyloIO event sequence into an application model using a hierarchical tree representation. In addition the usage of JPhyloIO factories and file filters in demonstrated.
- Detailed description
- Browse source codes of this demo
- SVN checkout:
https://secure.bioinfweb.info/Code/svn/JPhyloIO/trunk/demo/info.bioinfweb.jphyloio.demo.tree - Download executable demo files
Metadata demo
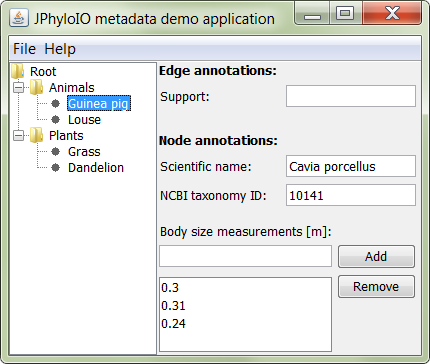
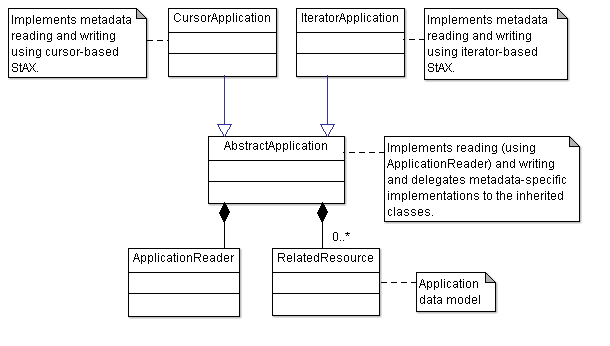
Shows the priciples of reading and writing metadata from and to different formats using JPhyloIO.
- Detailed description
- Browse source codes of this demo
- SVN checkout:
https://secure.bioinfweb.info/Code/svn/JPhyloIO/trunk/demo/info.bioinfweb.jphyloio.demo.metadata - Download executable demo files
XML Metadata demo
Shows the priciples of reading and writing XML representations of literal metadata elements using JPhyloIO.
- Detailed description
- Browse source codes of this demo
- SVN checkout:
https://secure.bioinfweb.info/Code/svn/JPhyloIO/trunk/demo/info.bioinfweb.jphyloio.demo.xmlmetadata - Download executable demo files
Usage examples in other projects
A number of available open-source tools make use of JPhyloIO. If you are looking for more advanced examples on how to use the functionality of the library, you can have a look at the following source codes. If you are new to JPhyloIO we recommend to have a look at the example applications listed above first.
TreeGraph 2
In its current version the phylogenetic tree editor TreeGraph 2 makes use of JPhyloIO to import trees including their annotations from NeXML. (Future versions of TreeGraph will make use of the full functionality by using JPhyloIO for reading and writing NeXML, PhyloXML, Nexus and Newick. This is currently not implemented, since the metadata model of TreeGraph will first be refactored to support RDF annotations.)
Taxonomic Editor of the EDIT Platform
The Taxonomic EDITor models alpha-taxonomic workflows and currently makes use of JPhyloIO to export single read alignments to all supported formats. An application-specific implementation of MatrixDataAdapter was used for that. (Future versions will also include an import function.)
LibrAlign
LibrAlign is a GUI library providing components for displaying and editing multiple sequence alignments and their attached raw and metadata. It makes use of JPhyloIO in reading and writing alignments and their metadata in a separate I/O module. Since JPhyloIO events are shared by separate model instances in LibrAlign, it makes use of the push-parsing tool classes.