JPhyloIO - A Java library for event-based reading and writing of different phylogenetic file formats through a common interface
JPhyloIO is an open source Java library for reading and writing phylogenetic file formats. The main aim is to provide access to various formats using a single interface, while being independent of the concrete application data model, to achieve maximal flexibility. It supports event based reading and writing of the following alignment and tree formats:
- NeXML including its different types of metadata
- Nexus (including the
TAXA,DATA,CHARACTERS,UNALIGNED,TREESandSETSblocks, as well as theMIXEDdata type extension defined by MrBayes). In addition a Nexus API is offered that allows application developers to easily add support for additional (custom) blocks and commands. - PhyloXML
- FASTA (including support for FASTA comments and column indices)
- Newick tree format
- Phylip
- Extended Phylip
- MEGA (including different types character set definitions, reading only)
- PDE (the format of the alignment editor PhyDE, reading only)
- XTG (the format of the phylogenetic tree editor TreeGraph 2, reading only)
Application developers are able to implement format-independent data processing by including event based readers from JPhyloIO via the abstract strategy pattern. All readers in JPhyloIO are designed to deal with large amounts of data (alignments with many and/or very long sequences, large trees) without using a great amount of resources (CPU or RAM).
JPhyloIO is distributed under LGPL and can be downloaded or checked out from the subversion repository.
Citation
JPhyloIO has been published in BMC Bioinformatics:
Stöver BC, Wiechers S, Müller KF: JPhyloIO: A Java library for event-based reading and writing of different phylogenetic file formats through a common interface. BMC Bioinformatics 2019, 20:402
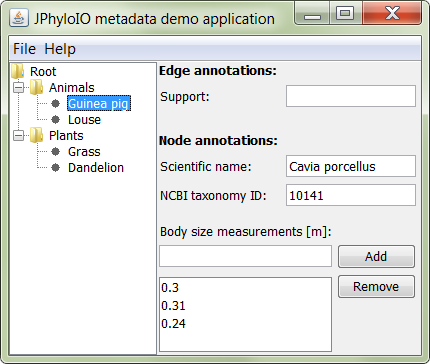
Look at the Demo Applications to quickly get started and see the possibilities of JPhyloIO.
Getting started
You can get an overview on how to use JPhyloIO in the documentation:
- Documentation overview
- Detailed JavaDoc
- Example applications showing how to use JPhyloIO
- JPhyloIO event lister shows how the libraries represents phylogenetic documents as an event stream
- Paper on JPhyloIO
If you have any questions regarding JPhyloIO that are not aswered in the documentation, feel free to ask a question in the
respective ResearchGate project or write an email to
support bioinfweb.info.
bioinfweb.info.
Current usage of JPhyloIO
JPhyloIO is currently used in the following software:
- The multiple sequence alignment GUI library LibrAlign
- The phylogenetic tree editor TreeGraph 2
- The Taxonomic Editor of the EDIT platform that manages alpha-taxonomic workflows and data
- AlignmentComparator used to compare alternative multiple sequence alignments of the same dataset
