JPhyloIO documentation
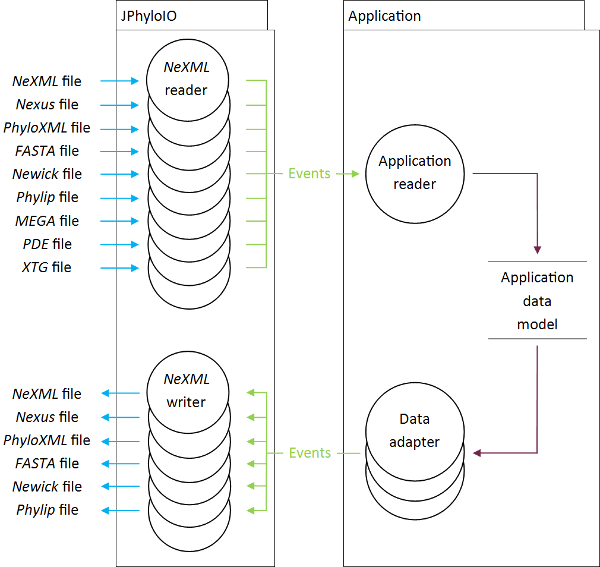
JPhyloIO is a Java library that allows bioinformatic application developers to support several phylogenetic file formats by just implementing one single reader and/or writer without detailed knowledge on the formats. Making the support of multiple formats easier should increase interoperability and foster the usage of more recently proposed powerful formats, such as NeXML. Our library allows to access nine phylogenetic file formats through one common interface, providing access to all features of each format (including complex metadata of NeXML and PhyloXML).
This page contains a brief introduction into the general architecture of JPhyloIO and is meant as the entry point to the full API documentation. For further details on how to use the library, we recommend to have a look at our example applications.
In addition to this documentation, the unit tests of JPhyloIO demonstrate how readers and writers are used and which events are generated from which files. The respective set of example files in different formats (used in the unit tests) shows the supported format variation of JPhyloIO.
If you have further questions on how to use the library, feel free to contact
stoever bioinfweb.info.
bioinfweb.info.
Contents of this page
- Event based reading
- Writing using data adapters
- Creating JPhyloIO reader and writer instances
- Handling metadata
Content of the documention
- Demo applications show how to use JPhyloIO in practice
- JPhyloIO event lister shows how the libraries represents phylogenetic documents as an event stream
- JavaDoc on the latest version
- JavaDocs on previous versions
Further reading
- The unit tests of JPhyloIO demonstrate how readers and writers are used and which events are generated from which files.
- A set of example files in different formats (used in the unit tests) that show the supported format variation of JPhyloIO
- A master thesis containing further information about the support of XML formats by JPhyloIO.
Event based reading
Documents read using JPhyloIO are translated to a stream of events that can be processed by the application in a memory efficient way. This allows developers full freedom in the design of their application data model, since no constraints are imposed by JPhyloIO. (The principle of phylogenetic document reading using JPhyloIO is similar to general XML reading using StAX. The difference is that JPhyloIO uses a larger set of events, specific for phylogenetics, as described below.)
To read data from the supported formats into the data model of an application based on JPhyloIO, one single reader class needs to be implemented in application code, that processes the event stream generated by the available document readers and this way acts as the mediator between the application data model and the event streams produced by JPhyloIO. (The advantage of implementing such a reader for JPhyloIO instead of implementing a reader directly for one specific format is that multiple formats are supported without additional work, increasing interoperability between applications.)
Currently the following reader implementations are available:
- NeXMLEventReader
- NexusEventReader
- PhyloXMLEventReader
- FASTAEventReader
- PhylipEventReader
- SequentialPhylipEventReader
- NewickEventReader
- MEGAEventReader
- XTGEventReader
- PDEEventReader
All readers implement the common interface
JPhyloIOEventReader
and can be exchanged via the strategy pattern. Instances
of format-specific readers (and writers) can be obtained using
JPhyloIOReaderWriterFactory
as described below. The documentation of
JPhyloIOEventReader
also contains the grammar defining how an event stream generated by any of its implementations looks like. Some events
have separated START and END events, that may enclose nested content, while others (which may not
contain nested content) only come in a single SOLE version. This is determined by their
EventTopologyType.
Important docs on event-based reading
Refer to the following resources for further details on event-based reading using JPhyloIO:
- JavaDoc of JPhyloIOEventReader (All format-specific readers implement this interface. Its documentation contains the event grammar defining how event streams in JPhyloIO look like.)
- EventContentType (Enumerates all types of JPhyloIO events. The documentation explains which event class is used for which event type.)
-
EventTopologyType
(Determines if an event type is represented by a
STARTandENDevent with possible nested content or by a singleSOLEevent.) - List of all event classes contained in JPhyloIO
- Example implementation of an application reader that processes events modeling an alignment (That class is part of the simple alignment demo application.)
- Example implementation of an application reader that processes events modeling a phylogenetic tree (That class is part of the tree demo application.)
- A conference poster on JPhyloIO providing an overview on the functionality
Writing using data adapters
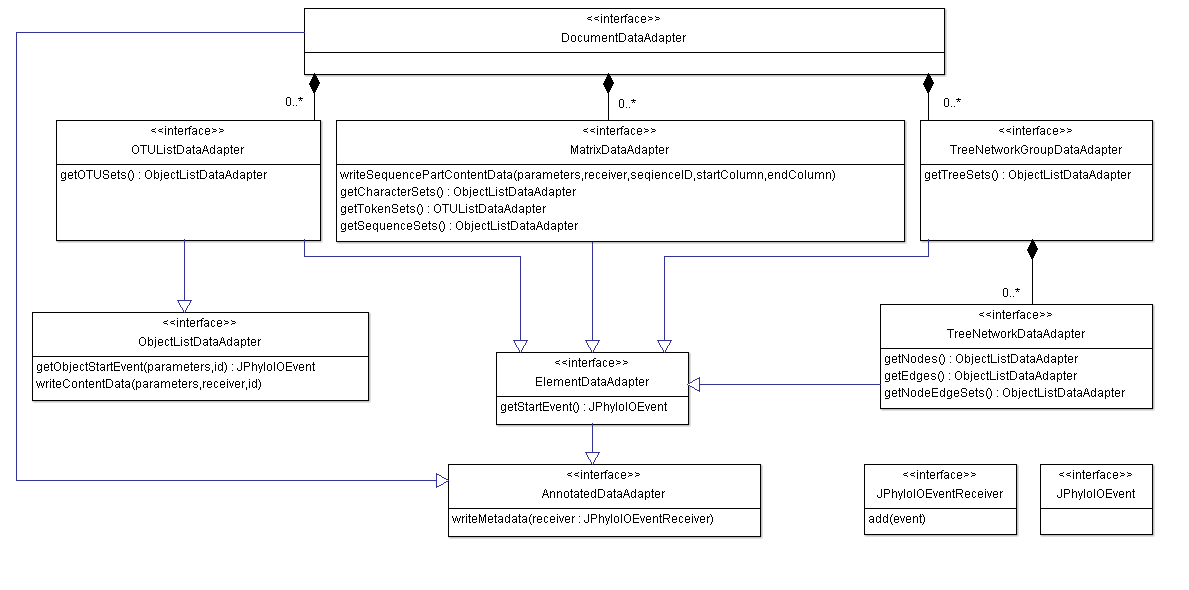
Data that is written using JPhyloIO is represented by the same event objects that are generated by the readers as described above. An event sequence must also be valid under the grammar to be found in the documentation of JPhyloIOEventReader. Since different formats require contents in different orders, JPhyloIO provides a set of data adapter interfaces that must be implemented by the application. Each adapter models a subsequence of events that corresponds to a certain grammar node. Figure 2 provides an overview on the different data adapters to be implemented. DocumentDataAdapter is the main adapter interface that provides access to all nested interfaces.
From top to bottom the object relation (indicated by aggregations) is shown, while the class hierarchy can be read from bottom to top. Note that not all but only exemplary methods are shown in each interface.
The
DocumentDataAdapter
is the main adapter that provides access to other adapters modelling OTU lists, matrices and phylogenetic trees or networks.
Not all application models will provide all these datatypes and therefore not need to implement all types of adapters.
The format specific writer classes in JPhyloIO can access the data either by event getter methods (e.g.
DocumentDataAdapter
MatrixDataAdapter.getSequenceStartEvent())
with an event ID as parameter or by writeXXX() methods (e.g.
MatrixDataAdapter.writeSequencePartContentData()),
which write a whole subsequence of the event stream to a special receiver object provided by the application. To simplify
the adapter implementation for application developers only frequently used events are provided by getter methods, while the
others can directly be written in a sequence by implementing an appropriate writer method. (Getter methods were introduced
for cases where random access to events with known IDs is frequently necessary for writers, to avoid requesting a whole
sequence, if only one event is needed. Providing some events by getter and some by writer methods in the data adapter model
is a compromise between ease of implementation and runtime performance.)
Some adapters share common functionality, which is modelled by common superinterfaces, such as AnnaotatedDataAdapter or ElementDataAdapter.
Note that not all adapters need to be implemented by each application, but only those relevant for the type of data that is handled by this program. In addition, JPhyloIO provides a set of abstract adapter implementations. that reduce the amount of programming work. In addition, so called store adapters. are provided that are complete implementations of the adapters and offer methods to add events that are than stored in the adapter instances to be provided to the writers. (Note that store adapters should only be used for simple cases with a small amount of data to be written. Otherwise application implementations of the necessary adapters that directly delegate to the application's data model classes should be preferred for memory efficiency.)
Important docs on data adapters and writing
Refer to the following resources for further details on writing documents using JPhyloIO:
- JavaDoc of JPhyloIOEventWriter (All format-specific writers implement this interface.)
- DocumentDataAdapter (The main data adapter interface that provides access to the whole document data, including nested adapter instances.)
- Overview on all available adapter interfaces
- Overview on all available abstract implementations of the adapter interfaces
- Overview on all store adapters
- Example implementation of a matrix data adapter (That class is part of the simple alignment demo application.)
- Example implementation of a tree/network data adapter that provides data on a phylogenetic tree together with an adapter implementation for nested node and branch lists (These classes are part of the tree demo application.)
- A conference poster on JPhyloIO providing an overview on the functionality
Creating JPhyloIO reader and writer instances
Format-specific event reader and writer instances provided by JPhyloIO can either be used directly by just creating instances of a concrete class or one of the tool methods of JPhyloIOReaderWriterFactory can be used. This class allows to guess the format of a file or stream and returns the according reader and is able to create instances of all available reader and writers.
Each of the different readers and writers make use of an instance of ReadWriteParameterMap that allows to specify certain parameters that influence the behavior of an I/O class. In addition this map is used by some instances to return additional information to the application. (Some writers e.g. add an instance of LabelEditingReporter to the map, which provides information to the application on how e.g. sequence labels had to be edited in order to match the restrictions of the target format.) The documentation of each reader and writer contains a list of supported parameters. In addition instances of JPhyloIOFormatInfo offer a method to programmatically check whether a parameter is supported by a reader or writer of a certain format.
Important docs on creating reader and writer instances
Refer to the following resources for further details on writing documents using JPhyloIO:
- JavaDoc of JPhyloIOReaderWriterFactory (Can be used to create instances and to guess the format of a stream or file.)
- ReadWriteParameterMap (The parameter map used to customize the behavior of readers and writers or to return additional data to the application.)
- ReadWriteParameterNames (A set of constants defining the names of all parameters supported by any of the reader and writer implementations available in JPhyloIO, including the documentation of each of these parameters.)
- JPhyloIOFormatInfo (Format-specific implementations of this class can be obtained using JPhyloIOReaderWriterFactory. They provide information of each format, including methods to programmatically determine which content and parameter is supported by each reader or writer.)
- The method Application.getFileChooser() in the tree demo application shows how file filters for all tree formats supported by JPhyloIO can be added to a file chooser dialog.
Handling metadata
The event grammar of JPhyloIO allows to nest metadata in most of the elements, which allows to annotate the whole document, sequences, tree nodes, OTUs and other elements. The general metadata concept allows to attach RDF annotations (a set of possibly nested resource and literal metadata elements) to each element.
Metadata is modeled by the following events:
- ResourceMetadataEvent indicates a resource annotation and allows to nest additional resource and literal metadata.
- LiteralMetadataEvent indicates a literal annotation and is followed by a sequence of nested content events.
- LiteralMetadataContentEvent represents literal metadata content. It may represent simple values in single events or in a sequence (e.g. for larger strings separated among multiple events) or an XML representation may be modeled by a sequence of such events. (See the class documentation for further details.)
Metadata in different formats
Although the full set of (possibly nested) RDF annotations (represented by respective event objects) may be passed to all JPhyloIO writers, it depends on the metadata support of the target format, if annotation events are ignored or not. NeXML supports the full set of annotations on every element and therefore also NeXMLEventWriter, but the others will ignore some events at certain positions and log according warnings. (Such warnings can be accessed using the logger instance specified in the parameter map passed to the event writer instance.) The documentations of the single writer classes contain information on the metadata support for each format. In addition the modeled metadata can also be checked programatically using JPhyloIOFormatInfo.getMetadataModeling(), which returns an instance of MetadataModeling which allows to determine which type of metadata is supported at the specified position. The metadata demo application shows how to read and write metadata in further detail.
Ways of reading and writing XML metadata
The usual way to read or write metadata would be to directly handle the respective JPhyloIO metadata content events described above in the application's reader or data adapter implementation. For XML literal metadata content, JPhyloIO offers to use cursor- and iterator-based StaX readers and writers as an alternative. This approach maybe more convenient on many cases and is especially usuful for applications that read and write XML annotations not only to phylogenetic file formats with JPhyloIO but in addition also to other (custom) XML files and therefore can use the same StaX code for both cases. The XML metadata demo application explains in detail how this is done.
Object translators
In addition to directly reading and writing LiteralMetadataContentEvents object translators exist in JPhyloIO. Implementations of the interface ObjectTranslator convert between a (possibly complex) Java object and its XML representation and optionally an alternative text representation. All JPhyloIO event readers and writers use an instance of ObjectTranslatorFactory that can be specified using their parameter map. By default a factory instance is used that is able to create object translators for the most XSD types, which are preimplemented in JPhyloIO. Custom object translator implementations can also be added to a factory and associated with a certain content type to be used by event readers and writers.
Note that JPhyloIO reader will automatically use all object translators available. That means that no literal content event sequences with single XML events will be generated, if a respective object translator is found. Instead a single content event containing the Java object generated by the translator will be fired.
Important docs on reading and writing metadata
Refer to the following resources for further details on reading and writing metadata in JPhyloIO:
- Metadata demo application showing the processing of different types of metadata and their support by different formats
- XML metadata demo application showing how to handle XML representations of literal metadata
- JavaDoc of MetaXMLStreamReader for cursor-based reading
- JavaDoc of MetaXMLStreamWriter for cursor-based writing
- JavaDoc of MetaXMLEventReader for iterator-based reading
- JavaDoc of MetaXMLEventWriter for iterator-based writing