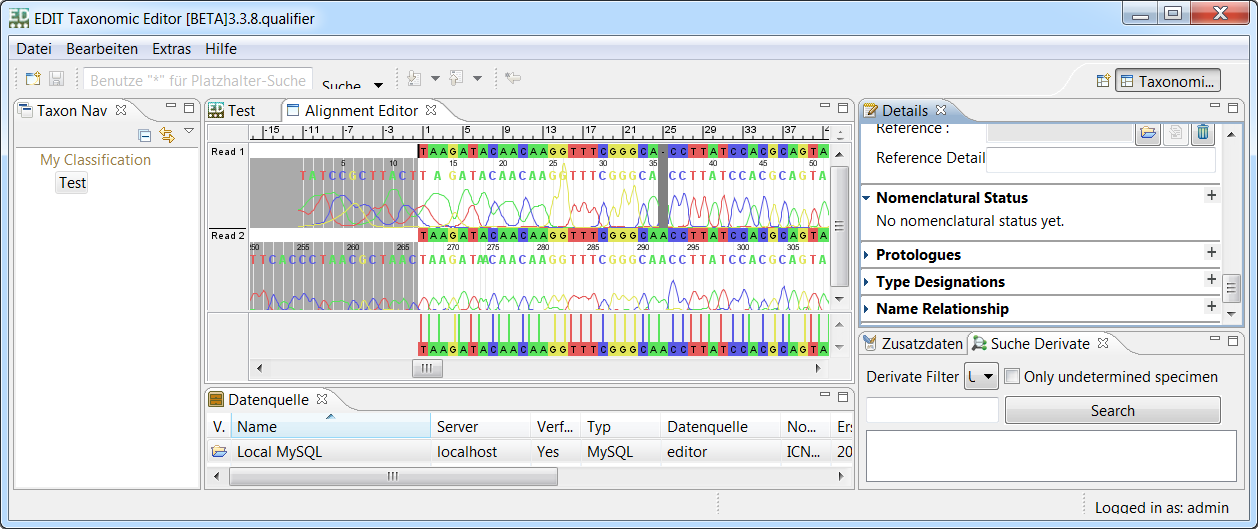
LibrAlign - A GUI library for displaying and editing multiple sequence alignments and attached data
LibrAlign is a Java library providing powerful GUI components for displaying and editing multiple sequence alignments. Data areas can be attached to sequences or whole alignments to display respective raw- and metadata.
Key features
- Flexible and powerful GUI components for displaying one or a set of MSAs together
- Data areas can be attached to each sequence or alignment. A set of predefined data areas is included and custom areas can easily be added using the API interfaces.
- All GUI components are based on TIC and can therefore be used both in Swing and SWT (including Eclipse RCP) applications.
- Reading and writing data and metadata from and to numerous alignment formats using JPhyloIO
- Different extensible data model implementations, including compressed sequence storage to handle large datasets
- Interoperability with BioJava
Getting started
Contact and feedback
If you have any question or feedback regarding LibrAlign feel free to contact
Ben Stöver
(stoever bioinfweb.info).
bioinfweb.info).
Software based on LibrAlign
Currently the following software uses LibrAlign components:
Future versions of the alignment editor PhyDE and tools for analyzing microstructural mutation patterns will also make use of LibrAlign.