JPhyloIO Metadata Demo
This example application demonstrates how to read and write metadata using JPhyloIO. It is an extension of the tree demo application. It is therefore necessary to understand the tree demo before proceeding here.
[Download executable demo files] [Browse source codes]
Overview
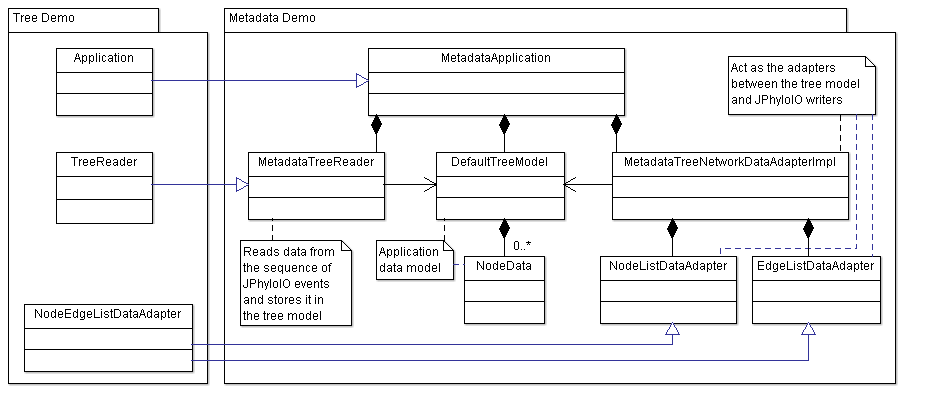
Since this demo application extends the functionality of the tree demo instead of reimplementing the general tree processing, it
extends classes from that demo and overwrites some methods that need to be adjusted to read and write metadata. As shown in
figure 1 the main class
MetadataApplication
extends
Application
and overwrites the methods readTree() and writeTree() as well as the getter methods providing
application name, version and URL.
To read a tree MetadataTreeReader is used, which is inherited from TreeReader as implemented in the tree demo. The writing of metadata attached to tree nodes and edges is implemented in adapters.NodeListDataAdapter and adapters.EdgeListDataAdapter, which are both inherited from tree demo class NodeEdgeListDataAdapter.
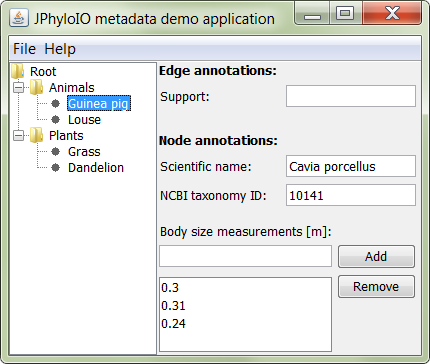
In addition to the topology read by the tree demo application, this demo program also models specific metadata attached to nodes and branches. Nodes carry two literal metadata string values (the scientific name and the NCBI taxonomy ID of the taxon) which are grouped by a parent resource metadata element modeling the taxonomy information of that node as a whole. Furthermore they may carry a list of size measurements of individuals of that species, which is stored in a single literal metadata element. Edges carry a numeric support value.
As shown in figure 2 all this metadata of a node and its afferent edge can be edited in the GUI after selecting that node. To store this metadata in formats that support it, necessary predicates identifying each metadata element modeled by this application are defined in the interface IOConstants. (Note that existing established ontologies should generally be used if available, but to keep this demo application more simple, we use this simple application specific example ontology.)
The following different examples of metadata shall demonstrate which type of annotations can be represented in which format and how a maximum format-independence can be achieved. Since NeXML offers the most comprehensive metadata model, all data can be represented there, while the other formats support only a subset of this.
Simple values attached to edges (the support value example)
The first metadata example in this application is a numeric support value attached to edge of a phylogenetic tree. (Note that it makes a difference if a value is attached to a node or an edge, e.g. if a tree is rerooted. The GUI of this demo application allows to edit the support value of the afferent edge of the selected node.) This simple numeric annotation can be represented in all tree formats that are supported by JPhyloIO and no format-specific implementations for reading or writing are necessary.
Writing
The support value is written to a file in the writeContentData() method of
EdgeListDataAdapter.
The easiest way to do that is to use the following tool method of
JPhyloIOWritingUtils:
JPhyloIOWritingUtils.writeSimpleLiteralMetadata(receiver, id + DEFAULT_META_ID_PREFIX + "Support", null,
PREDICATE_HAS_SUPPORT, DATA_TYPE_DOUBLE, support, null);
You can refer to the JavaDoc of
JPhyloIOWritingUtils.writeSimpleLiteralMetadata()
for details on all parameters. Important to mention
here is the event ID parameter. As shown above it is a concatenation of the edge ID (id), a meta ID prefix
(DEFAULT_META_ID_PREFIX as defined in
ReadWriteConstants)
and the string literal "Support". Such IDs need to be unique within a written document and they must be identical
for each event, even if the method writeContentData() of
EdgeListDataAdapter
is called multiple times, which is achieved by the implementation here. (Note that some writers will call this method multiple
times when writing a single document and do not work properly, if the ID of an event changes between the calls, e.g. because
IDs are generated dynamically by a counter.) The other parameters define the predicate for this type of metadata as
declared in
IOConstants,
the data type xs:double and the double value to be written (support).
Representation in the different formats
The support value annotation can be read and written by JPhyloIO from different formats, which use a different representation of such data. The data folder of the demo application in the subversion repository contains example outputs in all supported formats. You can also try this out yourself by loading a tree into the demo application and selecting Save as... from the main menu.
NeXML will model this annotation using its meta tag. See
NeXML.xml
for the full document.
<edge source="node0" target="node1" id="edge1" about="#edge1">
<meta id="edge1metaSupport" xsi:type="nex:LiteralMeta" property="a:isSupportedWith" datatype="xsd:double">99.0</meta>
</edge>
Newick and Nexus are able to represent simple edge annotations as hot comments inside the Newick string. See Newick.nwk and Nexus.nex for the full documents.
((Guinea_pig, Louse)Animals[&][&isSupportedWith=99.0], (Grass, Dandelion)Plants)Root;
The node annotations (described below) have been left out here. Note that the empty hot comment ([&]) preceding
the support value hot comment is necessary, since the first hot comment is considered as a node annotation and the second
as an edge annotation and no node attachments are present on the node Animals. The empty hot comment could also be
omitted if a branch length would have been specified. Since Newick and Nexus use string keys instead of CURIE
predicates and there is no possibility to declare namespaces, only the local part of the URI (isSupportedWith)
is written to the document.
PhyloXML uses property tags to represent custom annotations, which are not directly modeled in special
tags. (Note that PhyloXML does offer the confidence tag especially for support values, which should
usually be used. To achieve this a PhyloXML-specific predicate could be used, but in order to keep this example
simple, we decided to write the support value in the standard way here to demonstrate how simple literal attachments are
treated in general by JPhyloIO for PhyloXML. The taxonomy annotation described below will show how to make use
of special annotation tags in PhyloXML.) See
PhyloXML.xml
for the full document.
<clade id_source="node1">
...
<property ref="a:isSupportedWith" datatype="xsd:double" applies_to="parent_branch">99.0</property>
...
</clade>
The ref attribute allows to specify a predicate similar to the property attribute of the
meta tag in NeXML. The datatype attributes of both formats are also similar, but
PhyloXML is more limited here, since it does not allow externally defined data types, but only a set of values
defined in its schema.
This is not a problem here, but we will see in the size measurement example below that some workarounds are necessary,
if the property tag shall be used for data types that are not supported by PhyloXML. The
applies_to attribute determines whether an annotation is attached to the node or the edge.
(Note that although the ID of the literal metadata start event (that is written in the call of
JPhyloIOWritingUtils.writeSimpleLiteralMetadata()) is not contained in the PhyloXML file (since there are
no such metadata IDs modeled in the format),
PhyloXMLEventWriter
will anyway need a document-wide unique event ID to function properly.)
Reading
MetadataTreeReader
overwrites the method readEdgeContents() inherited from
TreeReader
to implement writing the support value. Analogous to reading, writing is also implemented using the tool
readLiteralMetadataContentAsObject from
JPhyloIOReadingUtils.
JPhyloIOEvent event = reader.next();
while (reader.hasNextEvent() && !event.getType().getTopologyType().equals(EventTopologyType.END)) {
if (event.getType().getTopologyType().equals(EventTopologyType.START)) {
if (event.getType().getContentType().equals(EventContentType.LITERAL_META)) {
LiteralMetadataEvent literalEvent = event.asLiteralMetadataEvent();
// Load possible support value:
if (PREDICATE_HAS_SUPPORT.equals(literalEvent.getPredicate().getURI())) {
((NodeData)targetNode.getUserObject()).setSupport(
JPhyloIOReadingUtils.readLiteralMetadataContentAsObject(reader, Number.class).doubleValue());
}
else { // Skip all nested events and the end event if another literal metadata element is nested.
JPhyloIOReadingUtils.reachElementEnd(reader);
}
}
else { // Skip possible other event subsequences.
JPhyloIOReadingUtils.reachElementEnd(reader);
}
}
event = reader.next();
}
This tool method consumes the whole event sequence from the literal metadata start to the respective end event and ignores any possible comment events. If an application is interested in such comments or if it expects very large string values that are separated over multiple instances of LiteralMetadataContentEvent it should implement processing the event sequence directly instead of using this method. This method is also not applicable to XML content of literal metadata annotations.
The above code snippet taken from
MetadataTreeReader.readEdgeContents()
subsequently reads events from the underlying JPhyloIO reader until the edge end event is reached. Within that loop
it checks whether a literal metadata start event with the predicate PREDICATE_HAS_SUPPORT (declared in
IOConstants)
is encountered. If so, its content is read using
JPhyloIOReadingUtils.readLiteralMetadataContentAsObject()
and stored into the
NodeData
instance of the respective tree node (which is the application's data model).
Since we are expecting a double value here, we try to parse in object that implements
Number.
Alternatively we could directly specify Double.class as the class parameter, but this way the implementation is
more flexibly and can also read files declaring other numeric data types like
Float,
Integer
or
BigDecimal.
As mentioned above Newick and Nexus use string keys instead of URIs to label metadata. Therefore a LiteralMetadataEvent produced by NewickEventReader or NexusEventReader will only have a string representation as a predicate and not a QName. To be able to load simple annotations (stored support values in this case) from Newick or Nexus documents, the condition in the code snippet above needs to be extended:
if (PREDICATE_HAS_SUPPORT.equals(literalEvent.getPredicate().getURI()) || PREDICATE_HAS_SUPPORT.getLocalPart().equals(literalEvent.getPredicate().getStringRepresentation())) { ... }
The second condition accepts predicates that only have the local part of the URI as their string representations. This way such metadata can be read from Newick and Nexus as well, but care should be taken if predicates from different ontologies (under different namespaces) are combined. In such cases it may happen that the string keys are ambiguous.
Nested RDF-annotations (the taxonomy example)
After we saw how to read and write simple (e.g. numeric) literal metadata annotations in the last section, we will now have a look at how to handle more complex metadata. We chose taxonomic information about a tree node as an example and read and write the scientific name and the NCBI taxonomy ID of the terminal nodes of the tree. For this example, we chose to group these two string literal metadata elements under a resource metadata element that models the taxonomy information as a whole. These three elements will be connected by the following predicates (declared in IOConstants):
PREDICATE_HAS_TAXONOMYconnects the resource metadata element modeling the taxonomy information as a whole to a tree node.PREDICATE_HAS_SCIENTIFIC_NAMEconnects the taxonomy resource metadata subject with the literal metadata object that contains the scientific name as a string.PREDICATE_HAS_NCBI_IDconnects the taxonomy resource metadata subject with the literal metadata object that contains the NCBI taxonomy ID as a string.
The following code writes the taxonomy resource element with its two nested literal elements. It is taken from
NodeListDataAdapter.writeContentData().
receiver.add(new ResourceMetadataEvent(id + DEFAULT_META_ID_PREFIX + "Tax1", null, new URIOrStringIdentifier(null, PREDICATE_HAS_TAXONOMY), null, null)); if ((data.getTaxonomy().getNCBIID() != null) && !data.getTaxonomy().getNCBIID().isEmpty()) { JPhyloIOWritingUtils.writeSimpleLiteralMetadata(receiver, id + DEFAULT_META_ID_PREFIX + "Tax2", null, PREDICATE_HAS_NCBI_ID, DATA_TYPE_STRING, data.getTaxonomy().getNCBIID(), null); } if ((data.getTaxonomy().getScientificName() != null) && !data.getTaxonomy().getScientificName().isEmpty()) { JPhyloIOWritingUtils.writeSimpleLiteralMetadata(receiver, id + DEFAULT_META_ID_PREFIX + "Tax3", null, PREDICATE_HAS_SCIENTIFIC_NAME, DATA_TYPE_STRING, data.getTaxonomy().getScientificName(), null); } receiver.add(ConcreteJPhyloIOEvent.createEndEvent(EventContentType.RESOURCE_META));
The tool method writeSimpleLiteralMetadata() is used in the same way as in the section above and its two calls are
surrounded by the highlighted code that writes an instance of
ResourceMetadataEvent
and its respective end event. (In the
full file
you find additional code that writes an alternative event sequence with PhyloXML-specific predicates. This is only
necessary if special annotation tags available in PhyloXML shall be supported. It is explained
below.)
The NeXML output resulting from executing the code abobe will look like this:
<node id="node2" about="#node2" label="Guinea pig">
<meta id="node2metaTax1" xsi:type="nex:ResourceMeta" rel="a:hasTaxonomy">
<meta id="node2metaTax2" xsi:type="nex:LiteralMeta" property="a:hasNCBIID" datatype="xsd:string">10141</meta>
<meta id="node2metaTax3" xsi:type="nex:LiteralMeta" property="a:hasScientificName" datatype="xsd:string">Cavia porcellus</meta>
</meta>
...
</node>
Reading files with nested metadata elements is similar to reading other nested events (e.g. the nodes within the tree or
the sequences within an alignment) which was explained in the previous demos. In this case the method
readStandardTaxonomy() was added to
MetadataTreeReader
that is called when the parent taxonomy resource start event is encountered and reads its nested sequence. Reading the two
literal metadata elements within this method in done in the same way is it was to read the support value
above. (Note that the alternative method readPhyloXMLTaxonomy() exists
as well. It is only necessary to read data from the PhyloXML taxonomy tag and is explained below.)
Using PhyloXML specific annotations
Unlike NeXML, PhyloXML does not allow to represent custom nested annotations (e.g. using property
tags), but if offers a set of predefined annotations to be represented in special tags. For taxonomic information the
taxonomy tag exists, which has a number of child elements including a scientific_name and an
id tag.
To make use of this special tags,
NodeListDataAdapter
checks in writeContentData() if the target format is PhyloXML or not. If so, it writes a different event
sequence then for other formats using PhyloXML-specific predicates:
if (parameters.getObject(KEY_WRITER_INSTANCE, null, JPhyloIOEventWriter.class).getFormatID().equals( JPhyloIOFormatIDs.PHYLOXML_FORMAT_ID)) { receiver.add(new ResourceMetadataEvent(id + DEFAULT_META_ID_PREFIX + "Tax1", null, new URIOrStringIdentifier(null, PhyloXMLConstants.PREDICATE_TAXONOMY), null, null)); // Write NCBI taxonomy ID to JPhyloIO: if ((data.getTaxonomy().getNCBIID() != null) && !data.getTaxonomy().getNCBIID().isEmpty()) { receiver.add(new ResourceMetadataEvent(id + DEFAULT_META_ID_PREFIX + "Tax3", null, new URIOrStringIdentifier(null, PhyloXMLConstants.PREDICATE_TAXONOMY_ID), null, null)); JPhyloIOWritingUtils.writeSimpleLiteralMetadata(receiver, id + DEFAULT_META_ID_PREFIX + "Tax4", null, PhyloXMLConstants.PREDICATE_TAXONOMY_ID_ATTR_PROVIDER, DATA_TYPE_STRING, PHYLOXML_ID_PROVIDER_NCBI, null); JPhyloIOWritingUtils.writeSimpleLiteralMetadata(receiver, id + DEFAULT_META_ID_PREFIX + "Tax5", null, PhyloXMLConstants.PREDICATE_TAXONOMY_ID_VALUE, DATA_TYPE_STRING, data.getTaxonomy().getNCBIID(), null); receiver.add(ConcreteJPhyloIOEvent.createEndEvent(EventContentType.RESOURCE_META)); // Terminate the taxonomy ID resource metadata element. } // Write scientific name to JPhyloIO: if ((data.getTaxonomy().getScientificName() != null) && !data.getTaxonomy().getScientificName().isEmpty()) { JPhyloIOWritingUtils.writeSimpleLiteralMetadata(receiver, id + DEFAULT_META_ID_PREFIX + "Tax2", null, PhyloXMLConstants.PREDICATE_TAXONOMY_SCIENTIFIC_NAME, DATA_TYPE_STRING, data.getTaxonomy().getScientificName(), null); } } else { // Write events for other formats ... // See code snippet above. }
There is predicate constant declared in PhyloXMLConstants for every metadata tag of PhyloXML. To write such specialized metadata tags, metadata events with these predicates can be written as shown above. Parent tags of PhyloXML are modeled as ResourceMetadataEvents, while attributes and text within such tags are modeled as LiteralMetadataEvents nested within the resource metadata events. Note that the order of writing for the events must match the order that is defined in the PhyloXML schema. Refer to the documentation of PhyloXMLEventWriter for further details.
The above code produces the following output in PhyloXML:
<clade id_source="node2">
...
<taxonomy>
<id provider="ncbi_taxonomy">10141</id>
<scientific_name>Cavia porcellus</scientific_name>
</taxonomy>
...
</clade>
The following event sequence is produced by JPhyloIO from the PhyloXML snippet above:
-
Start event of the type
EventContentType.RESOURCE_METAwith the relPhyloXMLConstants.PREDICATE_TAXONOMY-
Start event of the type
EventContentType.RESOURCE_METAwith the relPhyloXMLConstants.PREDICATE_TAXONOMY_ID-
Start event of the type
EventContentType.LITERAL_METAwith the predicatePREDICATE_TAXONOMY_ID_ATTR_PROVIDER- Sole event
EventContentType.LITERAL_META_CONTENTwith the content "ncbi_taxonomy"
- Sole event
- End event of the type
EventContentType.LITERAL_META -
Start event of the type
EventContentType.LITERAL_METAwith the predicatePREDICATE_TAXONOMY_ID_VALUE- Sole event
EventContentType.LITERAL_META_CONTENTwith the content "10141"
- Sole event
- End event of the type
EventContentType.LITERAL_META
-
Start event of the type
- End event of the type
EventContentType.RESOURCE_META -
Start event of the type
EventContentType.LITERAL_METAwith the predicatePREDICATE_TAXONOMY_SCIENTIFIC_NAME- Sole event
EventContentType.LITERAL_META_CONTENTwith the content "Cavia porcellus"
- Sole event
- End event of the type
EventContentType.LITERAL_META
-
Start event of the type
- End event of the type
EventContentType.RESOURCE_META
Reading taxonomy information from PhyloXML is implemented in the methods readPhyloXMLTaxonomy()
and readPhyloXMLTaxonomyID() of
MetadataTreeReader,
which process the event stream modeling the content of one taxonomy tag as shown above. Since the contents
of the id tag of PhyloXML (the text value and the provider attribute) are nested under
another resource metadata element, the additional method readPhyloXMLTaxonomyID() for processing the events
nested under this resource metadata event was implemented.
All code explained in this section is only necessary, of PhyloXML-specfic tags shall be supported. If no such
format-specific implementations are made, the genus and species name would be written into property tags on the
top level and the information about the parent resource metadata element (as it is written to NeXML) would be lost (in
the same way as it is described for Newick and Nexus below).
Note that the parameter KEY_PHYLOXML_METADATA_TREATMENT can be used to customize the way how
PhyloXMLEventWriter
handles nested metadata if no PhyloXML-specific predicates are used.
PhyloXMLMetadataTreatment
enumerated the different options.
Nested annotations in Newick and Nexus
The Newick and Nexus formats do not support nesting annotations.
NewickEventWriter
and
NexusEventWriter
only write terminal metadata events and ignore all resource elements that contain nestes elements. Therefore the
hasTaxonomy root element gets lost when writing to these formats and the genus and species annotations are on
the top level:
((Guinea_pig[&hasGenus=Cavia, hasSpecies=porcellus], Louse[&hasGenus=Gyropus, hasSpecies=ovalis]) ... )Root;
Due to the shift of the genus and species annotation to the top level, this demo application will not be able to read these annotations again from Newick or Nexus, which is intended to show which formats support nested annotations and which don't. It would of course be possible though to adjust MetadataTreeReader in a way that it can also read the genus and species name from literal metadata events with respective string predicates on the top level. Such an specialized implementation is not done in this example to keep it from becoming too complex.
Lists of simple values as annotations (the size measurement example)
In this last example a list of double values shall be attached to tree nodes. It represents a list of size
measuments obtained from different individuals of the respective species. Special hot comments for Newick and
Nexus exist that allow attaching a list of strings or numeric values to a node or an edge. If a literal metadata
event with any implementation of
Iterable
as its object value is passed to a JPhyloIO writer as shown below, it will automatically be written as a list.
The list ↔ string conversion is implemented in
ListTranslator.
JPhyloIOWritingUtils.writeSimpleLiteralMetadata(receiver, id + DEFAULT_META_ID_PREFIX + "Sizes", null,
PREDICATE_HAS_SIZE_MEASUREMENTS, DATA_TYPE_SIMPLE_VALUE_LIST, data.getSizeMeasurements());
If the code above is executed, the result in Newick and Nexus will look like this:
((Guinea_pig[&hasGenus=Cavia, hasSpecies=porcellus, hasSizeMeasurements={0.3, 0.31, 0.24}], Louse[&hasGenus=Gyropus, hasSpecies=ovalis, hasSizeMeasurements={0.0012, 9.0E-4, 0.0011, 0.0013, 0.0012}]) ... )Root;
To achieve a maximal format-independence, JPhyloIO supports writing such lists also to XML formats (although
these would of course also allow to use a special XML representation for a list). To achieve this the data type
DATA_TYPE_SIMPLE_VALUE_LIST (declared in
ReadWriteConstants)
must be specified for such a
LiteralMetaEvent.
The output in NeXML would be the following:
<node id="node2" about="#node2" label="Guinea pig">
...
<meta id="node2metaSizes" xsi:type="nex:LiteralMeta" property="a:hasSizeMeasurements"
datatype="jpd:simpleValueList">{0.3, 0.31, 0.24}</meta>
</node>
PhyloXML-specific reading
The representation in PhyloXML looks like this:
<property ref="a:hasSizeMeasurements" datatype="xsd:string" applies_to="node">{0.3, 0.31, 0.24}</property>
As mentioned above, PhyloXML only allows to use a predefined set of data type declarations, which is why the
JPhyloIO-specific data type jpd:simpleValueList cannot be written out to a document.
PhyloXMLEventWriter
uses xsd:string instead. As a consequence
ListTranslator
is not used for interpreting the string representation of that list, but it will be modeled as string literal metadata instead.
As a workaround for this limitation of the PhyloXML format, this example application tests if the read object is an
instance of list and if not manually converts the string:
Object list = JPhyloIOReadingUtils.readLiteralMetadataContentAsObject(reader, Object.class); if (list instanceof List) { // This case is used when reading valid documents of all formats but PhyloXML. data.setSizeMeasurements((List<Double>)list); // If the document is invalid, the list would not necessarily contain only double values. This would have to checked in a real-world application to avoid exceptions. } else if (list instanceof String) { // This block is used when reading valid PhyloXML documents. try { data.setSizeMeasurements((List)ListTranslator.parseList((String)list)); } catch (UnsupportedOperationException | InvalidObjectSourceDataException e) {} }
The code snippet above is taken from MetadataTreeReader.