JPhyloIO Tree Demo
This example application shows how to read phylogenetic trees from the non-hierarchical tree representation of the JPhyloIO event sequence into an application using DefaultTreeModel as its hierarchical tree model. Additionally, the usage of JPhyloIOReaderWriterFactory and the provided file filters is demonstrated in a GUI application.
[Download executable demo files] [Browse source codes]
Overview
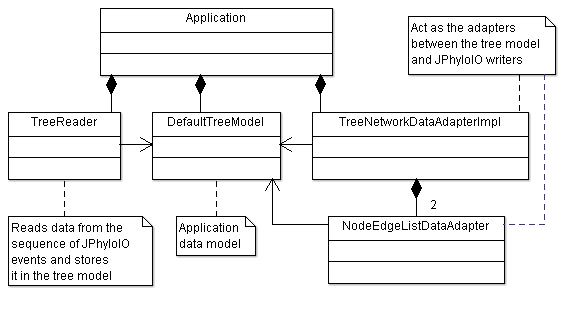
The main class of this example application is Application. It implements a Swing JFrame containing a JTree to display the loaded phylogenetic tree. It uses a JFileChooser to open and save trees in all formats supported by JPhyloIO as described below. As shown in figure 1 additional classes for reading and writing are part of the application and connect its business model with the event readers and writers of JPhyloIO.
Reading
The JPhyloIO event grammar models phylogenetic trees and networks the same way (except for the fact that networks may
not have root edges). (See grammar node TreeOrNetworkContent in the documentation of
JPhyloIOEventReader).
As a result, representations of phylogenetic trees are not hierarchical so that child nodes are nested between the
START and END events of their parent nodes, but instead all node definitions are on the same level,
independent of their position in the tree topology. Their connections are then defined by edge events that reference
a source and a target node by their ID (see
EdgeEvent).
This is closely related to the representation of trees in NeXML and is necessary to be able to
represent phylogenetic networks.
Applications not modeling networks and using a hierarchical tree representation in their business model (like DefaultTreeModel which is used in this example) need to translate the non-hierarchical event sequence into their hierarchical data structure. The class TreeReader in this example application implements this translation using the algorithm described in the documentation of TreeReader.readTree().
Writing
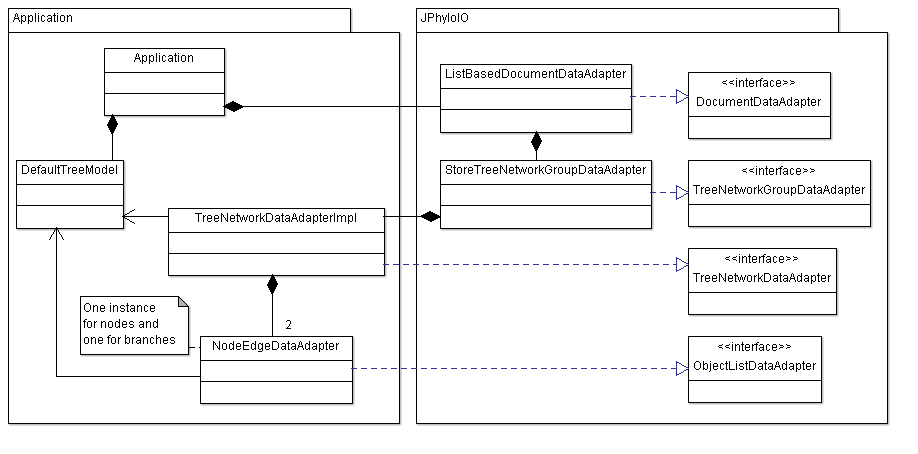
As mentioned in the simple alignment example, data adapters are used to connect the business model of an application to JPhyloIO writers. As shown in figure 3 this test application uses a set of data adapter implementations provided by JPhyloIO in combination with some custom implementations.
The JPhyloIO implementations used are ListBasedDocumentAdapter and StoreTreeNetworkGroupDataAdapter. No application specific implementations are necessary here since neither any other element of a document (an alignment or an OTU list) nor additional trees or networks are modeled by the application. The list based document adapter is just filled with a single tree/network group adapter, which is in turn filled with a single tree adapter in the respective method in Application.
In cases where data adapters need to provide access to data directly modeled in the application business model, custom
application data adapter implementations are necessary for efficient writing. This example application contains two
such implementations.
TreeNetworkDataAdapterImpl
implements
TreeNetworkDataAdapter
which provides a list of nodes and edges of the loaded tree. The node and edge lists are in turn instances of
NodeEdgeListDataAdapter
which implements
ObjectListDataAdapter.
NodeEdgeListDataAdapter
is designed to provide either all nodes or all edges from the tree model of the application as an object list.
It is an abstract class that keeps a list of all tree nodes to assign an index to each of them. This way each node
(and respective edge) can be referenced by an ID containing this index (e.g. node18). The creation of
concrete node or edge events is done in the abstract method createEvent(), which is implemented once
for nodes and once for edges by two anonymous classes in the
constructor of TreeNetworkDataAdapterImpl.
Using file filters and handling different formats
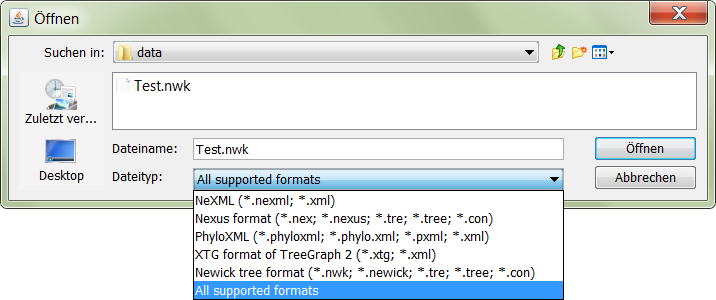
JPhyloIO offers file filters for all supported formats to be used e.g. with
JFileChooser.
These filters can be obtained from the respective
format info instance
which can in turn be obtained for each format using
JPhyloIOReaderWriterFactory.
Since this example application only models trees, the supported formats are filtered by their ability to support events
with the content type TREE in
Application.getFileChooser().