JPhyloIO XML Metadata Demo
In this example reading and writing of XML literal metadata is shown. It is assumed that you followed the previous examples, especially the application demonstrating how to read and write metadata in general. Furthermore you should be familiar with general event-based XML reading and writing in Java using the StAX API.
[Download executable demo files] [Browse source codes]
Overview
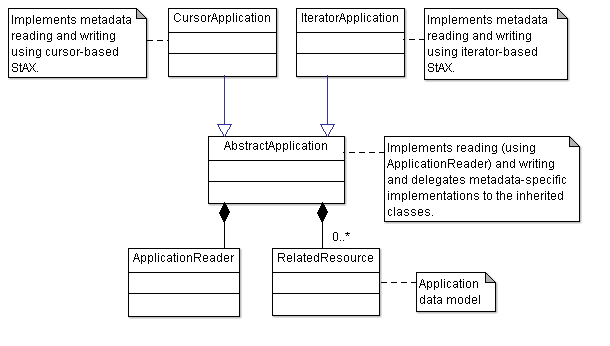
AbstractApplication
implements the basic behavior of both example applications and defines the abstract methods readMetadata()
and writeMetadata() which are used to read and write a single metadata element (a related reference as described
below). These methods are implemented by the two inherited classes
CursorApplication
and
IteratorApplication
which implement reading and writing of XML metadata using the cursor- and the iterator-based StAX approach.
The general architecture of the application is similar to that shown in the
simple alignment demo.
ApplicationReader
reads a document and calls the abstract method readMetadata() at the appropriate positions. Since the documents
written by the demo applications here do not contain any other data then the related references (metadata attached to the
document as a whole), there is not application specific adapter implementation necessary and an instance of
ApplicationReader
Modeled metadata
To keep this demo simple, we just read and write annotations on the document level here, although it would work the same way at any other position (e.g. under a tree node or a sequence). In addition to simple literal metadata annotations which have been introduced in the previous demo, JPhyloIO also supports complex literal values represented as XML. (Of course such data can only be written to XML formats like NeXML or PhyloXML. Writers for text formats like Nexus will ignore such metadata.)
The metadata modeled in this example is a set of resources related to the processed file. These relations are expressed as literal metadata annotations represented as XML:
<c:relatedReference c:type="WEBSITE"> <c:title>JPhyloIO</c:title> <c:url>http://bioinfweb.info/JPhyloIO/</c:url> </c:relatedReference>
In the example above, the related reference is the JPhyloIO website. The contents of such a literal value consist of the
title, the URL and the type of resource (a website in this case). Our example applications model such an annotation in the
class
RelatedResource
and its nested enum class
Type.
In this demo we will see how such an XML representation contained as metadata in a phylogenetic document can be read
into and written from these Java classes using JPhyloIO.
Different ways of reading and writing XML
JPhyloIO offers three different ways to read and write metadata:
- Direct processing of LiteralMetadataContentEvents containing XMLEvents as object values
- Iterator based StAX processing
- Cursor based StAX processing
In this example we will focus on StAX reading and writing and show both a cursor- and an iterator-based example. As mentioned above, JPhyloIO models any content of literal metadata annotations using one or more instances of LiteralMetadataContentEvent that contain a Java object and/or a string representation of the modeled metadata element. If the metadata element is represented as XML in a file it will be modeled as a sequence of LiteralMetadataContentEvents that contain a StAX XMLEvent as their object value each. The XML code of the related reference shown in the example above would be translated to the following sequence of events by a JPhyloIO reader (and can would be accepted by a writer in the same form):
-
Start event of the type
EventContentType.LITERAL_METAwith the sequence typeXML.-
Sole event of the type
EventContentType.LITERAL_META_CONTENTcontaining a StartElement instance as its object value that models the start tag<c:relatedReference c:type="WEBSITE">.-
Sole event of the type
EventContentType.LITERAL_META_CONTENTcontaining a StartElement instance as its object value that models the start tag<c:title>.- Sole event of the type
EventContentType.LITERAL_META_CONTENTcontaining a Characters instance as its object value that models the textJPhyloIO.
- Sole event of the type
- Sole event of the type
EventContentType.LITERAL_META_CONTENTcontaining a EndElement instance as its object value that models the end tag</c:title>. -
Sole event of the type
EventContentType.LITERAL_META_CONTENTcontaining a StartElement instance as its object value that models the start tag<c:url>.- Sole event of the type
EventContentType.LITERAL_META_CONTENTcontaining a Characters instance as its object value that models the texthttp://bioinfweb.info/JPhyloIO/.
- Sole event of the type
- Sole event of the type
EventContentType.LITERAL_META_CONTENTcontaining a EndElement instance as its object value that models the end tag</c:url>.
-
Sole event of the type
- Sole event of the type
EventContentType.LITERAL_META_CONTENTcontaining a EndElement instance as its object value that models the end tag</c:relatedReference>.
-
Sole event of the type
- End event of the type
EventContentType.LITERAL_META
Note that there may be additional literal metadata content events with XML character events as object values that model the whitespace between the tags, if the XML code contains the line breaks and indention as shown in the example above. The listing above does not contain these events for greater clarity.
Although it is possible for application code to process and generate such a sequence of JPhyloIO events directly, the library also offers implementations of the StAX reader and writer interfaces that can alternatively be used to read and write XML metadata. This is more convenient in many cases and possibly existing StAX code can be reused with JPhyloIO.
Cursor-based StAX processing of XML metadata
As mentioned before the basic functionality of reading and writing a document is implemented in the classes
AbstractApplication
and
ApplicationReader
which delegate XML reading and writing of a single "related reference" to classes inherited from
AbstractApplication.
CursorApplication
implements the respective abstract methods (readMetadata() and writeMetadata()) using the cursor
approach of StAX.
To read data this way an instance implementing XMLStreamReader needs to be obtained.
if (reader instanceof JPhyloIOXMLEventReader) {
XMLStreamReader xmlReader = ((JPhyloIOXMLEventReader)reader).createMetaXMLStreamReader();
... // Read data
}
JPhyloIOXMLEventReader
offers the method createMetaXMLStreamReader() that returns an implementation of
XMLStreamReader
that translates instances of
LiteralMetadataContentEvent
from its underlying JPhyloIO reader internally. It can only be called within the contents of a literal metadata
element, i.e. after a start event of the type
EventContentType.LITERAL_META
and before its respective end event. Calling the next() method of the
XMLStreamReader
will consume another event from the JPhyloIO reader by calling
JPhyloIOEventReader.next()
internally. The
XMLStreamReader
will not consume the LITERAL_META end event from the underlying reader, but will fire an
EndDocument
event after all XML content has been consumed. (Theoretically the application might even mix using both
next() methods in its code but that will usually not be beneficial.)
Since only JPhyloIOXMLEventReader and not JPhyloIOEventReader offers the methods to obtain StAX readers and writers, we need to check first, whether the used reader is an instance of JPhyloIOXMLEventReader. If not, the target format would be a text format (e.g. Nexus or FASTA) and events modeling XML metadata will anyway not be fired by such a reader, since its format cannot contain any XML annotations.
Reading the XML content can then be implemented as usual when using
StAX. In this example we use a
separate method that reads the contents of the parent relatedReference tag and some tool methods from the
bioinfweb.commons class
XMLUtils
are used.
To write data an instance of
XMLStreamWriter
needs to be obtained from the JPhyloIO event writer in the respective method of the data adapter implementation.
(Remember that the method writeMetadata() of the
DocumentDataAdapter
is delegated to
CursorApplication.writeMetadata()
in this example.)
if (parameters.get(ReadWriteParameterNames.KEY_WRITER_INSTANCE) instanceof JPhyloIOXMLEventWriter) { XMLStreamWriter writer = parameters.getObject(ReadWriteParameterNames.KEY_WRITER_INSTANCE, null, JPhyloIOXMLEventWriter.class).createMetaXMLStreamWriter(receiver); ... // Write data }
As shown in the previous example applications, writing data to JPhyloIO is achieved by the application by implementing
data adapters. Since this implementation can potentially be called by different instances of
JPhyloIOEventWriter,
the writer instance should be obtained from the provided parameter map instead of using some global application specific
field or property. Each implementation of
JPhyloIOEventWriter
will put a reference to itself into its parameter map under the key
ReadWriteParameterNames.KEY_WRITER_INSTANCE
and pass this map to every method of a data adapter it calls. Therefore the JPhyloIO writer instance can be obtained
as shown above. If that writer is an instance of
JPhyloIOXMLEventWriter
it will offer a method createMetaXMLStreamWriter() that returns an implementation of
XMLStreamWriter
that delegates to the underlying JPhyloIO event writer.
Writing the XML content can again be implemented as usual when using StAX.
Iterator-based StAX processing of XML metadata
In addition to the cursor-based approach, JPhyloIO also allows to use iterator-based StAX. According instances of XMLEventReader and XMLEventWriter can also be obtained from respective methods of JPhyloIOXMLEventReader and JPhyloIOXMLEventWriter as shown in the code examples below.
Reading:
if (reader instanceof JPhyloIOXMLEventReader) {
XMLEventReader xmlReader = ((JPhyloIOXMLEventReader)reader).createMetaXMLEventReader();
... // Read data
}
Writing:
if (parameters.get(ReadWriteParameterNames.KEY_WRITER_INSTANCE) instanceof JPhyloIOXMLEventWriter) {
XMLEventWriter writer = parameters.getObject(ReadWriteParameterNames.KEY_WRITER_INSTANCE, null,
JPhyloIOXMLEventWriter.class).createMetaXMLEventWriter(receiver);
... // Write data
}
The above code snippets are taken from IteratorApplication which implements iterator-based StAX XML reading and writing in the same way as CursorApplication did for the cursor-based approach.